As here we are working with many data, so the chances for data to be incorrect, duplicate or mislabeled. Data Cleaning is the process of changing or removing the incorrect, duplicate, corrupted or incomplete data in a dataset.
There are no precise steps available for the process of data cleaning because it can vary from dataset to dataset.
Having clean data will ultimately increase overall productivity and permit the very best quality information in our decision-making. In order to get accurate and valid visualization data cleaning plays a very important role.
So in this case, we will begin by dropping the unnecessary columns that are Province/State, Lat and Long. We will drop these columns using pandas.We can drop the column in many ways:
Dropping column using drop() function
confirmed_df=confirmed_df.drop(['Province/State'],axis=1)
recovered_df=recovered_df.drop(['Province/State'],axis=1)
death_df=death_df.drop(['Province/State'],axis=1)
Dropping column using iloc[ ] function
confirmed_df=confirmed_df.drop(confirmed_df.iloc[:,1:3],axis=1)
recovered_df=recovered_df.drop(recovered_df.iloc[:,1:3],axis=1)
death_df=death_df.drop(death_df.iloc[:,1:3],axis=1)
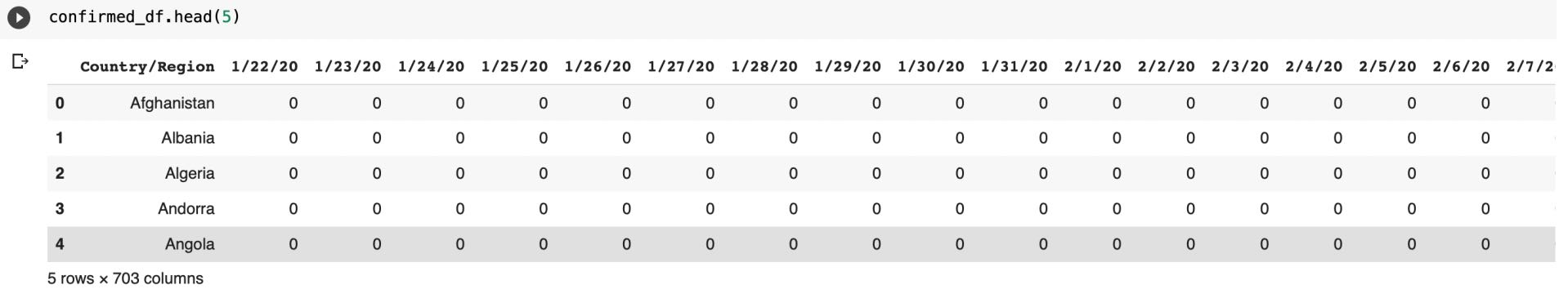
Then we will rename some columns
confirmed_df = confirmed_df.rename(columns={"Country/Region": "Country"})
recovered_df = recovered_df.rename(columns={"Country/Region": "Country"})
death_df = death_df.rename(columns={"Country/Region": "Country"})Then we will group all these data by its country name and then transpose it to get data in better format.
confirmed_df=confirmed_df.groupby(['Country']).aggregate(np.sum).T
recovered_df=recovered_df.groupby(['Country']).aggregate(np.sum).T
death_df=death_df.groupby(['Country']).aggregate(np.sum).TSetting the index name to Date to make data more accurate.
confirmed_df.index.name='Date'
confirmed_df=confirmed_df.reset_index()
recovered_df.index.name='Date'
recovered_df=recovered_df.reset_index()
death_df.index.name='Date'
death_df=death_df.reset_index()Now we will unpivot the datasets using melt() function because it is present in wide format and we need data in long format because this will make it easy to analyze.
confirmed_cases_df=confirmed_df.melt(id_vars="Date")
recovered_cases_df=recovered_df.melt(id_vars="Date")
death_cases_df=death_df.melt(id_vars="Date")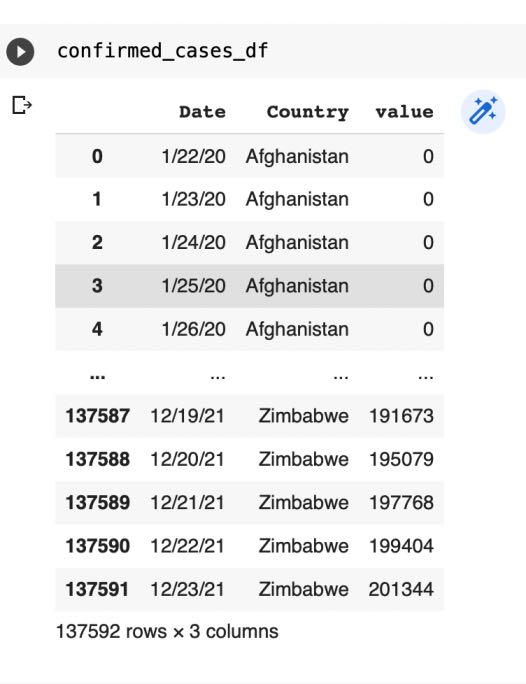
Now we will rename the values column
confirmed_cases_df=confirmed_cases_df.rename(columns={"value":"Confirmed Cases"})
recovered_cases_df=recovered_cases_df.rename(columns={"value":"Recovered Cases"})
death_cases_df=death_cases_df.rename(columns={"value":"Death Cases"})
As we have observed that the name of "values" column has been changed to "Confirmed Cases" but we can also see that date column is present in object format so we will change its data type.
confirmed_cases_df['Date']=pd.to_datetime(confirmed_cases_df['Date'])
recovered_cases_df['Date']=pd.to_datetime(recovered_cases_df['Date'])
death_cases_df['Date']=pd.to_datetime(death_cases_df['Date'])For Confirmed Cases
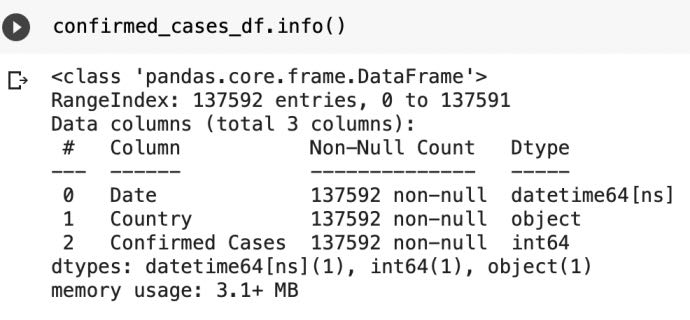
For Death Cases
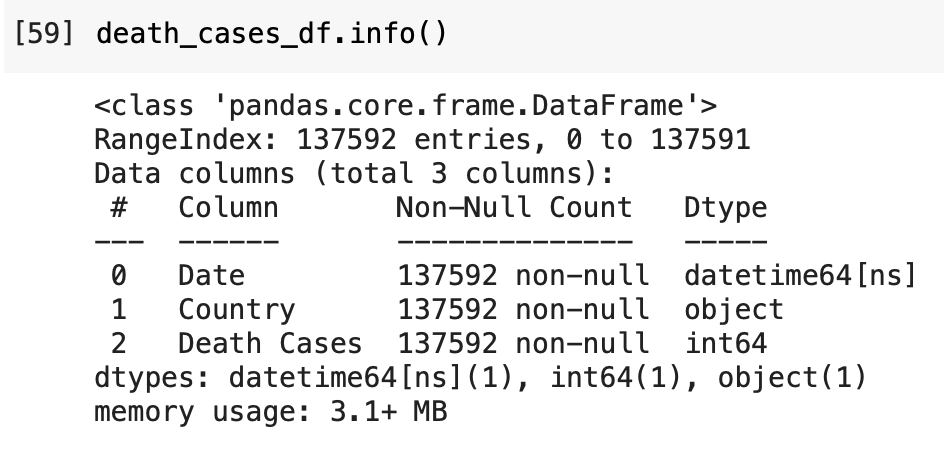
For Recovered Cases
- Log in to post comments